-Search query
-Search result
Showing all 21 items for (author: goldsmith & ja)

EMDB-41158: 
CS2it1p2_F7K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-41180: 
Global reconstruction for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab
Method: single particle / : Goldsmith JG, McLellan JS

EMDB-26738: 
Integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab
Method: single particle / : Goldsmith JA, McLellan JS
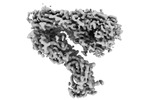
EMDB-26739: 
Integrin alphaM/beta2 ectodomain
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-27122: 
Global refinement for integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-27123: 
Tailpiece local refinement for integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-27124: 
Acylation domain local refinement for integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab
Method: single particle / : Goldsmith JA, McLellan JS
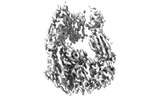
EMDB-27125: 
Global refinement for integrin alphaM/beta2 ectodomain
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-27126: 
Tailpiece local refinement for integrin alphaM/beta2 ectodomain
Method: single particle / : Goldsmith JA, McLellan JS

PDB-7usl: 
Integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab
Method: single particle / : Goldsmith JA, McLellan JS

PDB-7usm: 
Integrin alphaM/beta2 ectodomain
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-25072: 
Fab22 bound to MERS-CoV Spike
Method: single particle / : Hsieh CL, McLellan JS

EMDB-25073: 
Structure of Fab22 complexed SARS-CoV-2 spike
Method: single particle / : Hsieh CL, McLellan JS

EMDB-23156: 
SARS-CoV 2 Spike Protein bound to LY-CoV555
Method: single particle / : Goldsmith JA, McLellan JS

PDB-7l3n: 
SARS-CoV 2 Spike Protein bound to LY-CoV555
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-22221: 
SARS-CoV-2 HexaPro S One RBD up
Method: single particle / : Wrapp D, Hsieh CL, Goldsmith JA, McLellan JS

EMDB-22222: 
SARS-CoV-2 HexaPro S Two RBD up
Method: single particle / : Wrapp D, Hsieh CL, Goldsmith JA, McLellan JS

PDB-6xkl: 
SARS-CoV-2 HexaPro S One RBD up
Method: single particle / : Wrapp D, Hsieh CL, Goldsmith JA, McLellan JS

EMDB-21374: 
2019-nCoV spike glycoprotein with C3 symmetry imposed
Method: single particle / : Wrapp D, Wang N, McLellan JS

EMDB-21375: 
Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up
Method: single particle / : Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C, Abiona O, Graham BS, McLellan JS

PDB-6vsb: 
Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up
Method: single particle / : Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C, Abiona O, Graham BS, McLellan JS
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
